Overview of workshops
Core programme overview
Beginner
• Introduction to Shell for bioinformatics
• Introduction to R for bioinformatics
• RNA-seq Data Analysis
• Genomic Data Carpentry (Aotearoa edition) coming soon…
Intermediate
• Intermediate Shell for bioinformatics
• Intermediate R for bioinformatics
• Introduction to Bash Scripting and HPC Job Scheduler
Specialised
• Visualisation with ggplot2
• Reproducibility with Git and Quarto
Ancillary programme overview
Other specialised workshops
• Single-cell RNAseq Data Analysis
• Reproducible Bioinformatics Workflows with Nextflow and nf-core
• Microbial genome assembly with short reads
• Long read genome assembly
• Constructing Pangenome Graphs
• Outlier analysis
• Scaling gene regulatory network simulations
• Introduction to Software Containers
Got a workshop topic suggestion? Let us know!
Core programme
Introductory level
Our series of introductory workshops are designed to take you from absolute beginner to self-driving learner. Each of these workshops can be done as a standalone crash course in a particular topic, and they can also be strung together to provide a strong foundation in bioinformatics principals and practices.
Introduction to Shell for bioinformatics

Learn the fundamentals of working with the Command Line Interface (CLI). Shell is a program that allows you to interact with the command line. Familiarity with the shell will allow you to access remote servers, automate tasks, and use a wide range of tools that are unavailable on a Graphical User Interface (GUI).
During this workshop you will learn:
- The importance of the shell.
- How to navigate files and directories.
- How to create, view and modify files.
- Pipes, redirection, and scripts, which will allow you to automate your workflow.
Prerequisites: We assume the learner has no prior experience with the tools covered in the workshop. However, learners are expected to have some familiarity with biological concepts.
Format: Taught over one day (10am - 4pm).
View the full workshop material here: Introduction to Shell
Introduction to R for bioinformatics

Get started with R, a highly popular programming language in the fields of biology and statistics. R is world-renowned for producing high-quality, publication-ready figures and tables.
Note that this workshop is a pre-requisite for the RNA-seq Data Analysis workshop and intermediate R workshops.
Some of the topics covered in the workshop are:
- An introduction to R and RStudio.
- R basics: The R language, reading data into R, storing data as objects.
- R packages.
- Publication-quality data presentation using
ggplot2. - Where to get more help when you are ready to do more.
Prerequisites: We assume the learner has no prior experience with the tools covered in the workshop. However, learners are expected to have some familiarity with biological concepts.
Format: Taught over one day (10am - 4pm).
View the full workshop material here: Introduction to the R Programming Language
RNA-seq Data Analysis
Get started with analysing RNA-seq datasets, identifying differentially expressed genes and highlighting impacted biological processes.
Some of the topics covered in the workshop are:
- Quality assessment
- Trimming and filtering
- Mapping and read counts
- Differential expression analysis
- Over-representation analysis
Prerequisites: This is a beginner-friendly workshop and no prior experience in analysing RNA-seq data is required. However, we assume the learner has familiarity with basic transcriptomic and biological concepts, in particular that they know what sequencing libraries are and have some familiarity with the data format (i.e, know what a FASTA/FASTQ file is). Familiarity with beginner-level R and bash is also assumed. If you would like a refresher on R, see Introduction to R. If you would like a refresher on bash, see Introduction to Shell.
Format: Taught over two half days (9am - 1pm).
View the full workshop material here: RNA-seq Data Analysis
Intermediate level
Our intermediate level workshops are designed to build on skills learned in our introductory level, to enable you to more efficiently analyse your data and streamline your workflow.
Intermediate Shell for Bioinformatics

Shell overview, downloading and verifying data, inspecting and manipulating text data with Unix tools, automating file-processing.
This includes:
- An overview of the Shell, UNIX and Linux.
- Downloading data from a remote source and checking data integrity.
- Recap navigating files and directories, and commands used in routine tasks.
- Inspecting and manipulating data (the
head,tail,grep,sedandawkcommands).
- Automating file processing.
- Challenges: solve example molecular biology problems using shell scripts.
Prerequisites: Comfortable using bash / shell at a beginner level (have completed Introduction to Shell or have equivalent experience).
Format: Taught over one day (10am - 4pm).
View the full workshop material here: Intermediate Shell for Bioinformatics
Intermediate R for bioinformatics

Advance your skills with R! You will learn to complete R tasks with fewer lines of code, scale your analyses, and write readable code.
Some of the topics covered in the workshop are:
- Introduction to relational data and the join function.
- Working with regular expressions and functions from the stringr package.
- Writing custom functions, working with conditional statements.
- ‘Defensive programming’.
- Iterations - for loops, and
map_*()functions.
- The importance of data structure in R.
Prerequisites: Comfortable using R / R Studio at a beginner level (have completed Introduction to R or have equivalent experience)
Format: Taught over two half days (9am - 1pm).
View the full workshop material here: Intermediate R
Introduction to Bash Scripting and HPC Job Scheduler
Write your own bash scripts for data analysis and working on an HPC (high performance computing) environment.
Some of the topics covered in the workshop are:
- Designing a variant calling workflow.
- Automating a workflow.
- An introduction to HPC.
- Working with job scheduler.
Prerequisites: Comfortable using bash / shell at a beginner level (have completed Introduction to Shell or have equivalent experience). Being comfortable with a genomic analysis pipeline at a beginner level (e.g., have completed RNA-seq Data Analysis or similar workflow) will be useful but is not required.
We assume learners have familiarity with genomic concepts, but do not specifically need to be working on variant calling workflows to learn the basics of bash scripting and job scheduling in this workshop.
Format: Taught over one day (10am - 4pm).
View the full workshop material here: Introduction to Bash Scripting and HPC Scheduler
Specialised
These workshops are designed to address specific concepts or teach specific workflows. Generally these workshops require some beginner knowledge of R or shell.
Visualisation with ggplot2

Visualisation is more than just the code used to make a plot.
The aim for this workshop is to showcase the full process of visualising data. This includes some basic exploratory analysis, some minor data transformation, and then thinking about the visual story. Finally, a fully realised visualisation will be created.
This workshop is split into four parts:
- Basic
ggplot2format and showcase of what can be done.
- Some data transformations and tidying, viz for exploratory analysis.
- Group walkthrough of creating a visualization of example data.
- Working on your own data: plan, transform data, visualize.
Prerequisites: Comfortable using R / R Studio at a beginner level (have completed Introduction to R or have equivalent experience). Some familiarity with ggplot2 will be useful but not required.
Format: Taught over one day (10am - 4pm).
View the full workshop material here: From Start to Finish: Visualising Your Data
Reproducibility with Git and Quarto

Good research is about more than just doing the analysis; it’s also about making your analysis reproducible, collaborative, and easy to share with your supervisor, your colleagues, or the wider scientific community.
This workshop will teach you how to:
- Use Git and GitHub to confidently host and manage your own code, and collaborate with others.
- Tell the story of your analysis with clear, self-contained Quarto documents.
- Create polished HTML outputs with well-documented code and embedded results.
- Share your work with the world by publishing it as a website via GitHub Pages.
Prerequisites: This is an intermediate level workshop. We assume the learner has no prior experience with the tools covered in the workshop, but you must have attended our Introduction to R workshop and Introduction to Shell workshop, or have equivalent experience. You should be comfortable navigating the file system on your own computer using commands such as cd, pwd and ls and be comfortable with basic R/RStudio syntax (e.g., know what functions, objects and vectors are, and have used RStudio before).
Set-up: This workshop is run locally on your own computer. Before the workshop, you must install R, RStudio and Git, and make a GitHub account. The full set-up instructions can be found here.
Format: Taught over two half days (9:30am - 2:30pm).
View the full workshop material here: Reproducibility with Git and Quarto
Ancillary programme
Our ancillary programme are our highly specialised topics, generally at an intermediate to advanced level. These workshops are typically scheduled based on expression of interest and availability of topic-specialist instructors.
Single-cell RNA-seq data analysis
Learn the skills and tools required for the analysis of single-cell RNA-seq data (scRNA-seq data) in R.
This workshop covers:
- Alignment and feature counting with Cell Ranger (briefly).
- QC and exploratory analysis.
- Normalisation.
- Sctransform: Variant Stabilising transformation.
- Feature selection and dimensionality reduction.
- Batch correction and data set integration.
- Clustering.
- Identification of cluster marker genes.
- Differential gene expression analysis.
- Differential abundance.
Prerequisites: This is an advanced workshop which requires an intermediate level of R knowledge and experience. To participate, you must have completed Intermediate R or have equivalent experience.
Format: Taught over 4 half days (9am – 1pm).
View the full workshop material here: Analysis of single-cell RNA-seq data
Reproducible Bioinformatics with Nextflow and nf-core

Reproducible research is of the utmost importance. Nextflow is workflow management software that enables writing scalable and reproducible scientific workflows. It integrates software packages and environment management systems from environment modules to Docker, Singularity, and Conda.
In this workshop you will:
- Be introduced to Nextflow and execute an example pipeline.
- Be introduced to nf-core, an online repository of curated pipelines.
- Learn how to configure and customise an existing nf-core pipeline.
- Generate metrics and reports.
Prerequisites: Comfortable using command line / shell at a beginner-intermediate level.
Format: Taught over one day (10am - 4pm).
View the full workshop material here: Reproducible Bioinformatics Workflows with Nextflow and nf-core
Microbial genome assembly with short reads
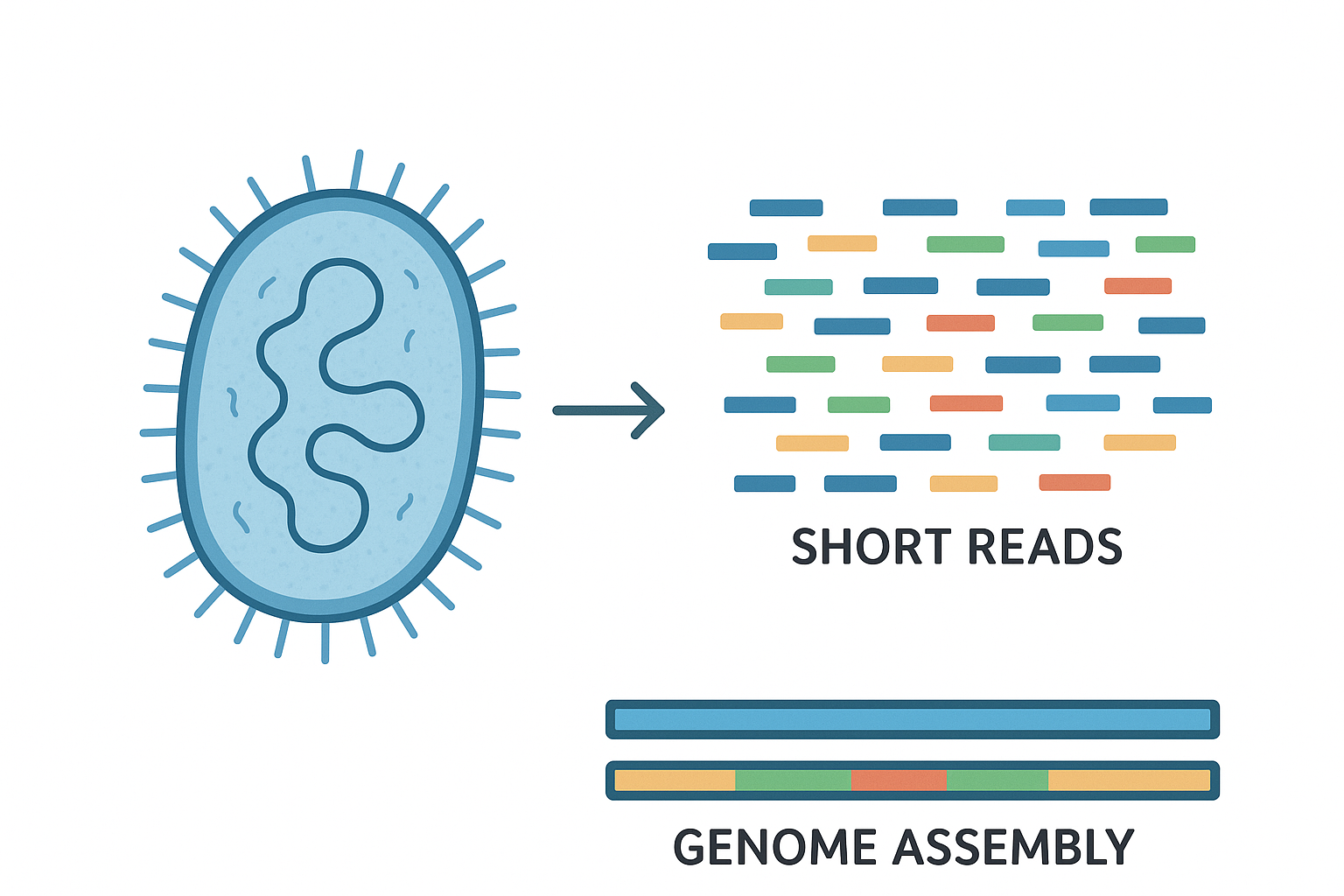
This workshop aims to provide a comprehensive understanding of microbial genome assembly using short-read sequencing data.
This workshop will cover:
- The principles of microbial genome assembly using short sequencing reads.
- Differences between de novo and reference-guided assembly approaches.
- Hands-on walkthrough of a genome assembly workflow.
- Key considerations such as sequencing read length, depth, and contamination.
- Genome annotation and visualisation techniques.
- Practical examples using Microcoleus cyanobacterial sequencing data.
Prerequisites: Some familiarity with the command line and basic R. Comfortable with: navigating files, using Slurm/HPC, command-line tools, and basic R.
Format: Taught over two half days (9am - 1pm).
View the full workshop material here: Microbial Genome Assembly with Short Reads
Long read genome assembly
This long read assembly workshop works through an entire genome assembly workflow including data QC, assembly, and assembly QC.
Some of the topics covered:
- Sequence data basics: HiFi and UltraLong read data specifics
- Quality Control (QC) of the data: cleaning, read length filtering. Overview of phasing.
- Assembly of a genome: Verkko and Hifiasm, comparison of approaches.
- Assembly QC: biological and technical assessments of the three Cs (contiguity, correctness, completeness).
- Contiguity using gfastats
- Correctness using Merqury
- Completeness using asmgene
- Assembly cleanup and genome annotation: contamination checks, Liftoff, MashMap, Minimap2.
- Phased assemblies: benefits and examples.
Prerequisites: Introductory knowledge of bash / command line (e.g., completed Introduction to Shell).
Format: Taught over one day (10am - 4pm).
View the full workshop material here: Long read assembly
Constructing Pangenome Graphs
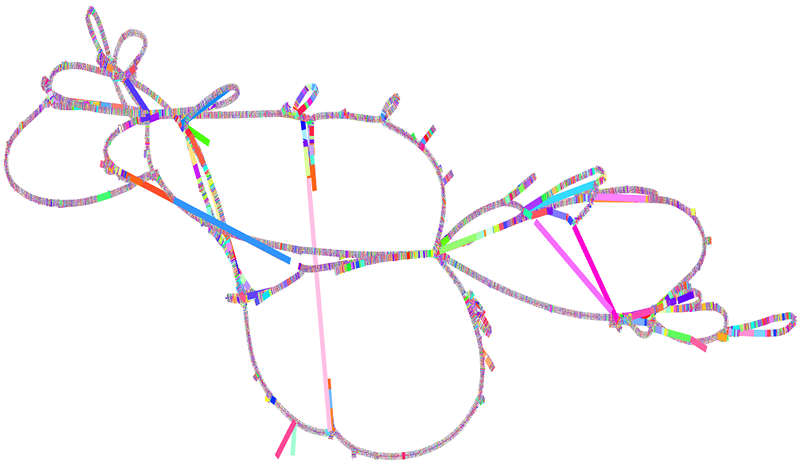
How to construct a pangenome graph using PGGB, including QC, variant extraction, and short-read mapping.
This workshop will include:
- Introduction to pangenome graphs.
- Setup guide for using the tools and data.
- Overview of the PGGB toolkit.
- Choosing parameters to construct a graph.
- QC, extracting variant data, mapping short reads.
Prerequisites: Familiarity with Shell. Able to navigate files/directories, use full vs relative paths, and use a command-line text editor.
Format: Taught over one day (10am - 4pm).
View the full workshop material here: Unlock the Power of Pangenome Graphs
Outlier Analysis
Identify genomic regions under selection using the outlier analysis method.
During this workshop:
- Download example genomic data or prepare your own.
- Use PCAdapt to identify outlier loci.
- Use VCFtools to identify outlier SNPs in population comparisons.
- Use Bayescan to identify outlier SNPs based on allele frequencies.
- Relate identified SNPs to phenotypic variation.
- Compare results of different methods and discuss findings.
Prerequisites: Familiarity with R and basic command line. Some knowledge of genomic concepts and selection.
Format: Taught over two full days (10am - 4pm).
View the full workshop material here: Outlier Analysis
Scaling Gene Regulatory Networks Simulations
Simulate gene regulatory networks using R and Julia.
This workshop will include:
- Why simulations are valuable in systems biology.
- What regulatory networks are and how to model them.
- Using the sismonr R package to simulate a small network.
- Introduction to HPC: architectures, batch systems, and Slurm.
- How to scale up simulations on HPC via profiling and optimisation.
Prerequisites: Familiarity with bash and R; some HPC knowledge preferred. Basic molecular biology knowledge helpful.
Format: Taught over two full days (10am - 4pm).
View the full workshop material here: Scaling Gene Regulatory Networks Simulations
Introduction to Software Containers

This workshop introduces Apptainer, showing how to run a simple container and build your own, including running parallel scientific workloads on HPC clusters.
View the full workshop material here: Introduction to Software Containers
